中国全科医学 ›› 2023, Vol. 26 ›› Issue (08): 939-950.DOI: 10.12114/j.issn.1007-9572.2022.0487
所属专题: 心血管最新文章合辑
柴晏1, 赵玉青2, 郭旭男1, 王东英1, 边云飞3,*( )
)
收稿日期:2022-07-06
修回日期:2022-08-28
出版日期:2023-03-15
发布日期:2022-11-30
通讯作者:
边云飞
基金资助:
CHAI Yan1, ZHAO Yuqing2, GUO Xunan1, WANG Dongying1, BIAN Yunfei3,*( )
)
Received:2022-07-06
Revised:2022-08-28
Published:2023-03-15
Online:2022-11-30
Contact:
BIAN Yunfei
摘要: 背景 心血管疾病(CVD)是常见病和多发病,患病率和死亡率呈快速上升趋势。动脉粥样硬化(AS)是缺血性CVD的病理基础,研究表明心外膜脂肪组织(EAT)通过分泌外泌体(EXO)和生物活性物质促进AS进展,但其作用机制仍需进一步研究。 目的 通过生物信息学方法挖掘冠状动脉粥样硬化性心脏病(CAD)患者EAT中的关键基因,探讨免疫细胞浸润情况,联合CAD患者EXO间差异表达基因(DEGs)推测EAT来源EXO间DEGs并进行验证,从细胞及分子水平上探讨EAT在CAD疾病过程中的作用机制。 方法 从基因表达综合数据库(GEO)中下载关于EAT的数据集GSE64554、GSE120774,根据临床信息将EAT的测序数据分为CAD组和健康对照组,使用R语言及相关软件包进行生物信息学分析。首先使用R语言筛选CAD组与健康对照组EAT间DEGs,并进行GO富集分析和KEGG通路富集分析,构建蛋白质-蛋白质相互作用(PPI)网络,评估所选基因的生物学功能及可能参与其调控的转录因子。构建GSE64554数据集中EAT的加权基因共表达网络(WGCNA),获取同CAD表型相关的基因模块,将所获EAT间DEGs与模块内hub基因取交集获得关键基因,采用Cibersort反卷积算法对EAT组织的免疫细胞浸润情况进行分析。通过exoRbase数据库获取CAD组与健康对照组血液EXO间DEGs,CAD组和健康对照组EAT间DEGs与EXO间DEGs取交集作为CAD的诊断、治疗标志物,收集临床样本通过实时荧光定量PCR(qRT-PCR)对其进行验证。对所选基因进行GO/KEGG富集分析和Metascape富集分析。 结果 共筛选出CAD组与健康对照组EAT间DEGs 1 511个,其中表达上调的基因956个,表达下调的基因555个。通过对CAD表型相关的模块内枢纽基因与EAT间DEGs取交集获得EAT在CAD发生、发展中的关键基因DDX47、FEM1C、NOL11、SRP54、ABI1、PATL1、BNIP2、C1orf159、CHCHD4。免疫细胞浸润分析显示,CAD组EAT中幼稚CD4+ T细胞表达丰度升高而静息树突细胞表达丰度减低(P<0.05)。筛选获得CAD EXO间DEGs 1 658个,其中表达上调的基因278个,表达下调的基因1 380个,EAT间DEGs与EXO间DEGs取交集,共获得129个基因,选取表达丰度较高的BPI、BIRC5、CXCL12、RNASE1、F2R作为CAD患者潜在诊断、治疗标志物,经qRT-PCR验证结果显示,CAD组与健康对照组比较BPI、BIRC5、CXCL12和RNASE1的mRNA水平升高(P<0.05),F2RmRNA水平下降(P<0.05)。GO/KEGG富集分析显示EAT间DEGs主要富集于细胞质基质、MHC蛋白复合物、RNA降解、抗原加工和呈递等,构建PPI网络,通过Cytoscape软件CytoHubba插件MCC算法获得连接度最高的基因RPS27A。Metascape富集分析显示DEGs主要富集于细胞对DNA损伤刺激反应、RNA代谢、调节细胞对压力的反应、适应性免疫系统等,TRRUST数据库预测CIITA转录因子可能参与了EAT间DEGs的调控。 结论 (1)EAT可能通过促炎和免疫途径参与CAD的发生和发展,DDX47、FEM1C、NOL11、SRP54、ABI1、PATL1、BNIP2、C1orf159、CHCHD4、RPS27A可能作为关键基因并发挥重要作用。(2)CAD患者EAT中幼稚CD4+ T细胞表达丰度升高而静息树突细胞表达丰度减低。(3)BPI、BIRC5、CXCL12、RNASE1、F2R可能由EAT分泌并可作为CAD的诊断、治疗标志物。
| 引物名称 | 引物序列 | 产物长度(bp) | 退火温度(℃) |
|---|---|---|---|
| H-BPI-S | GAATCAACTATGGTCTGGTGGCA | 106 | 60.62 |
| H-BPI-A | AAGGGAGGTGGATTGTGGTG | ||
| H-BIRC5-S | CGCATCTCTACATTCAAGAACTGG | 182 | 59.73 |
| H-BIRC5-A | GGTTTCCTTTGCATGGGGT | ||
| H-CXCL12-S | TGAGCTACAGATGCCCATGC | 137 | 60.18 |
| H-CXCL12-A | GCACACTTGTCTGTTGTTGTTCTTC | ||
| H-RNASE1-S | GCTCCTTCTGCTTGTCCTGAT | 149 | 60.07 |
| H-RNASE1-A | TCATCATTTGGTTACAGTAGGTGGA | ||
| H-F2R-S | CAGTGATTGGCAGTTTGGGTCT | 270 | 61.08 |
| H-F2R-A | TGACAGGTAGTGATGTTGAGCCC | ||
| H-ACTIN-S | CACCCAGCACAATGAAGATCAAGT | 317 | 61.60 |
| H-ACTIN-A | CCAGTTTTTAAATCCTGAGTCAAGC |
表1 PCR引物
Table 1 Primer sequences for qRT-PCR
| 引物名称 | 引物序列 | 产物长度(bp) | 退火温度(℃) |
|---|---|---|---|
| H-BPI-S | GAATCAACTATGGTCTGGTGGCA | 106 | 60.62 |
| H-BPI-A | AAGGGAGGTGGATTGTGGTG | ||
| H-BIRC5-S | CGCATCTCTACATTCAAGAACTGG | 182 | 59.73 |
| H-BIRC5-A | GGTTTCCTTTGCATGGGGT | ||
| H-CXCL12-S | TGAGCTACAGATGCCCATGC | 137 | 60.18 |
| H-CXCL12-A | GCACACTTGTCTGTTGTTGTTCTTC | ||
| H-RNASE1-S | GCTCCTTCTGCTTGTCCTGAT | 149 | 60.07 |
| H-RNASE1-A | TCATCATTTGGTTACAGTAGGTGGA | ||
| H-F2R-S | CAGTGATTGGCAGTTTGGGTCT | 270 | 61.08 |
| H-F2R-A | TGACAGGTAGTGATGTTGAGCCC | ||
| H-ACTIN-S | CACCCAGCACAATGAAGATCAAGT | 317 | 61.60 |
| H-ACTIN-A | CCAGTTTTTAAATCCTGAGTCAAGC |
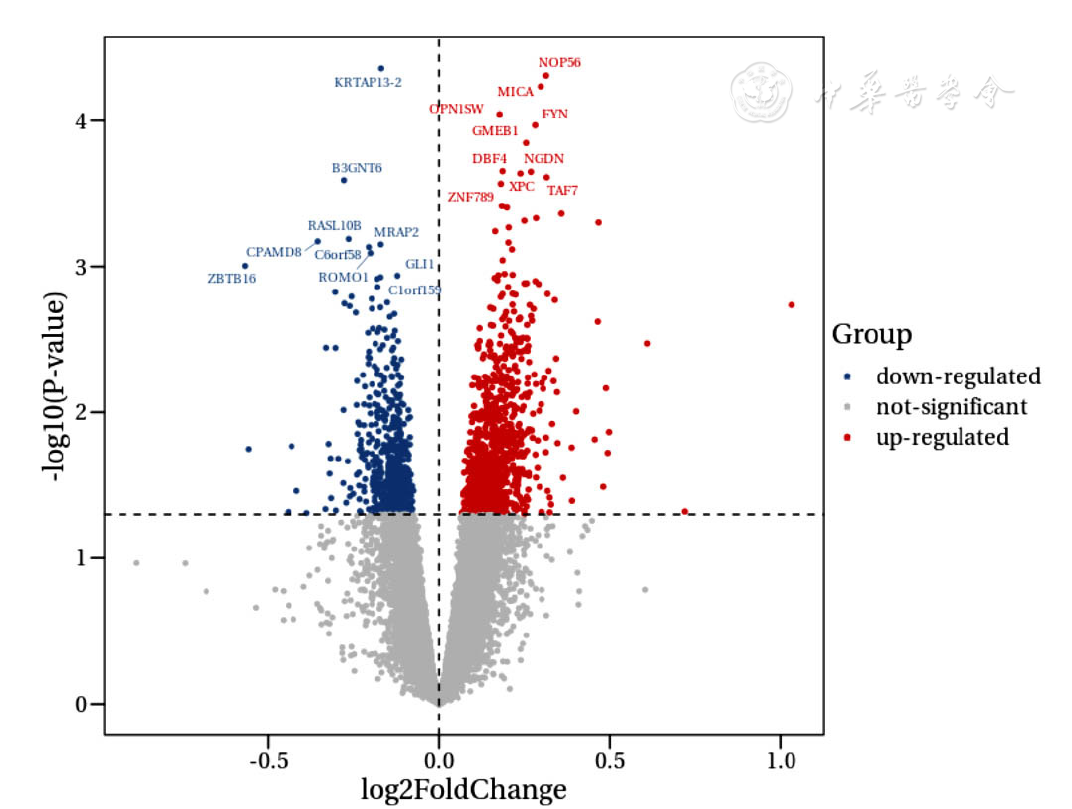
图1 CAD组与健康对照组EAT间DEGs火山图
Figure 1 Volcano plot of differentially expressed genes in epicardial adipose tissue between CAD group and healthy control group
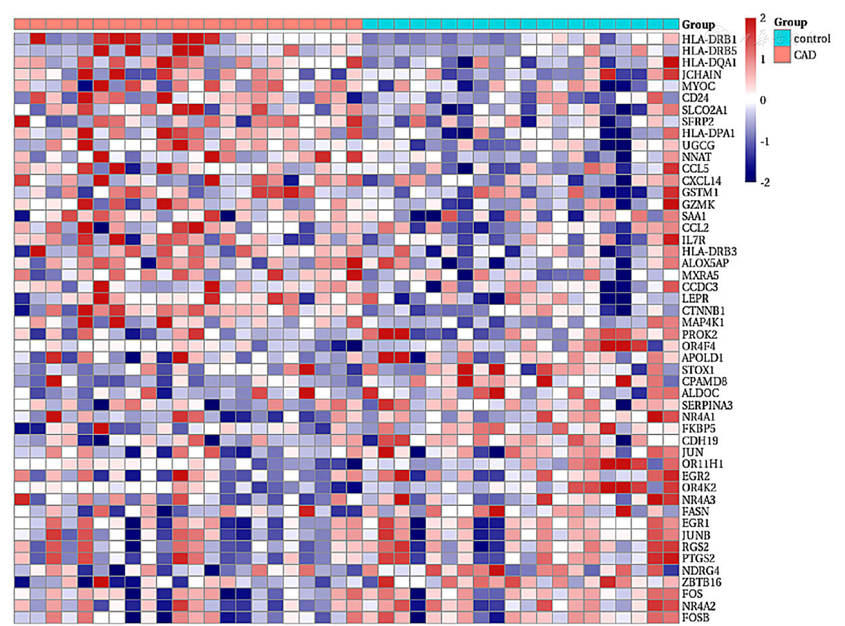
图2 CAD组与健康对照组EAT间DEGs热图注:CAD=冠状动脉粥样硬化性心脏病
Figure 2 Heatmap of differentially expressed genes in epicardial adipose tissue between CAD group and healthy control group
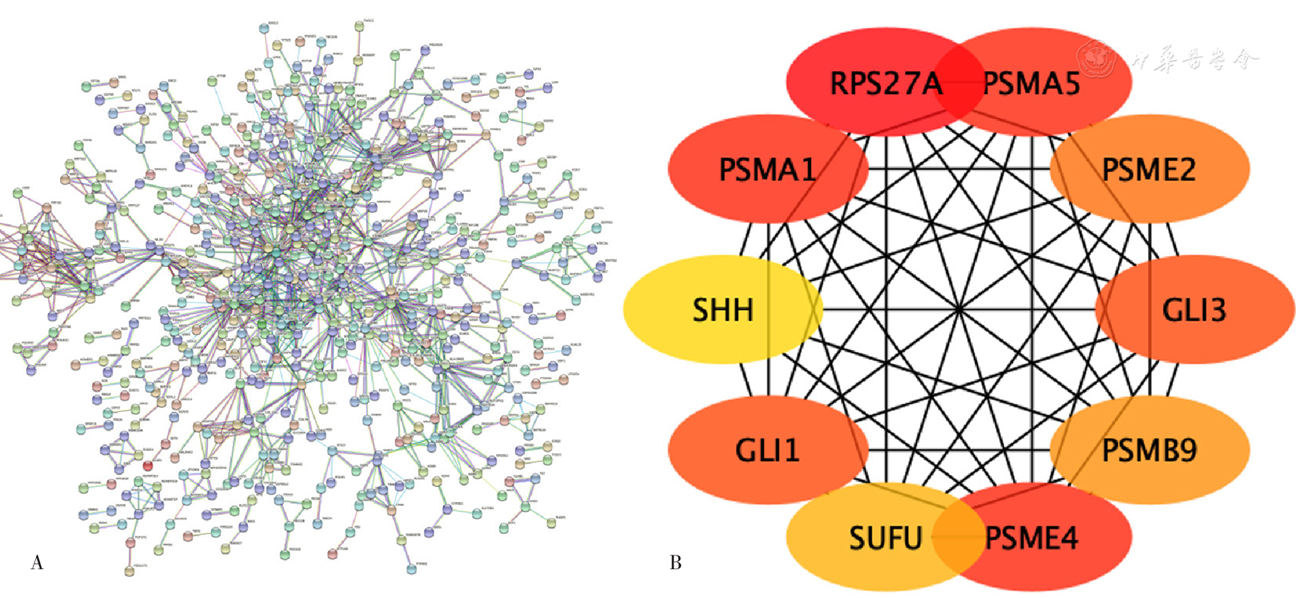
图5 EAT间DEGs的PPI网络与关键基因注:A为EAT间DEGs的PPI网络图,B为EAT间DEGs的关键基因图
Figure 5 Differentially expressed genes in epicardial adipose tissue and the key genes identified in the PPI network
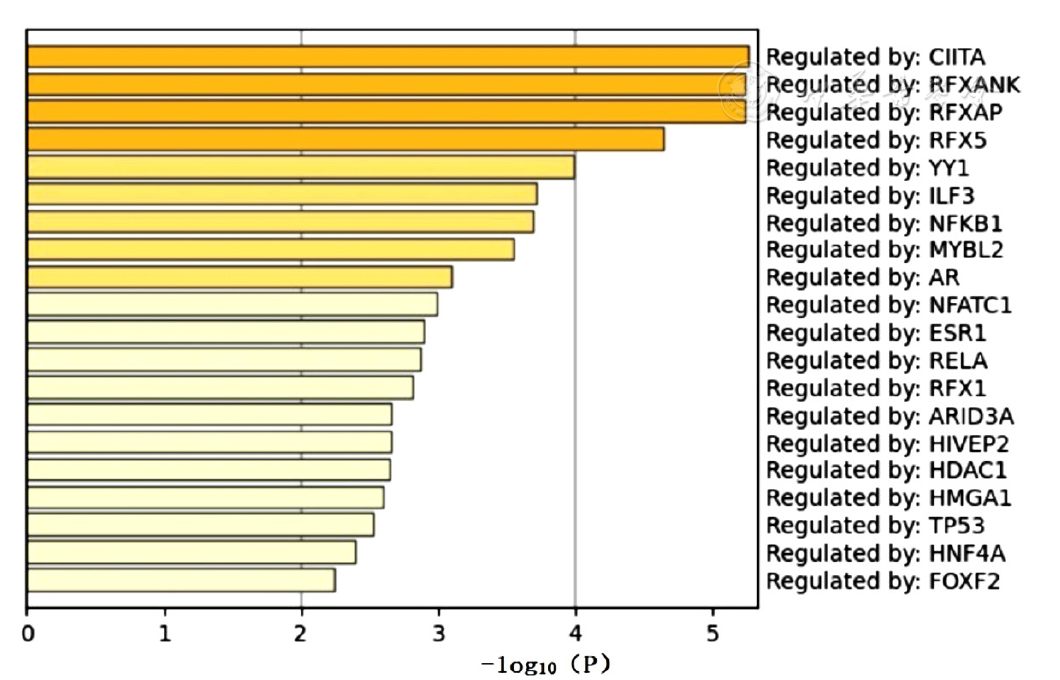
图7 EAT间DEGs的TRRUST数据库预测转录因子
Figure 7 TRRUST database-based analysis of differentially expressed genes in epicardial adipose tissue to predict transcription factors
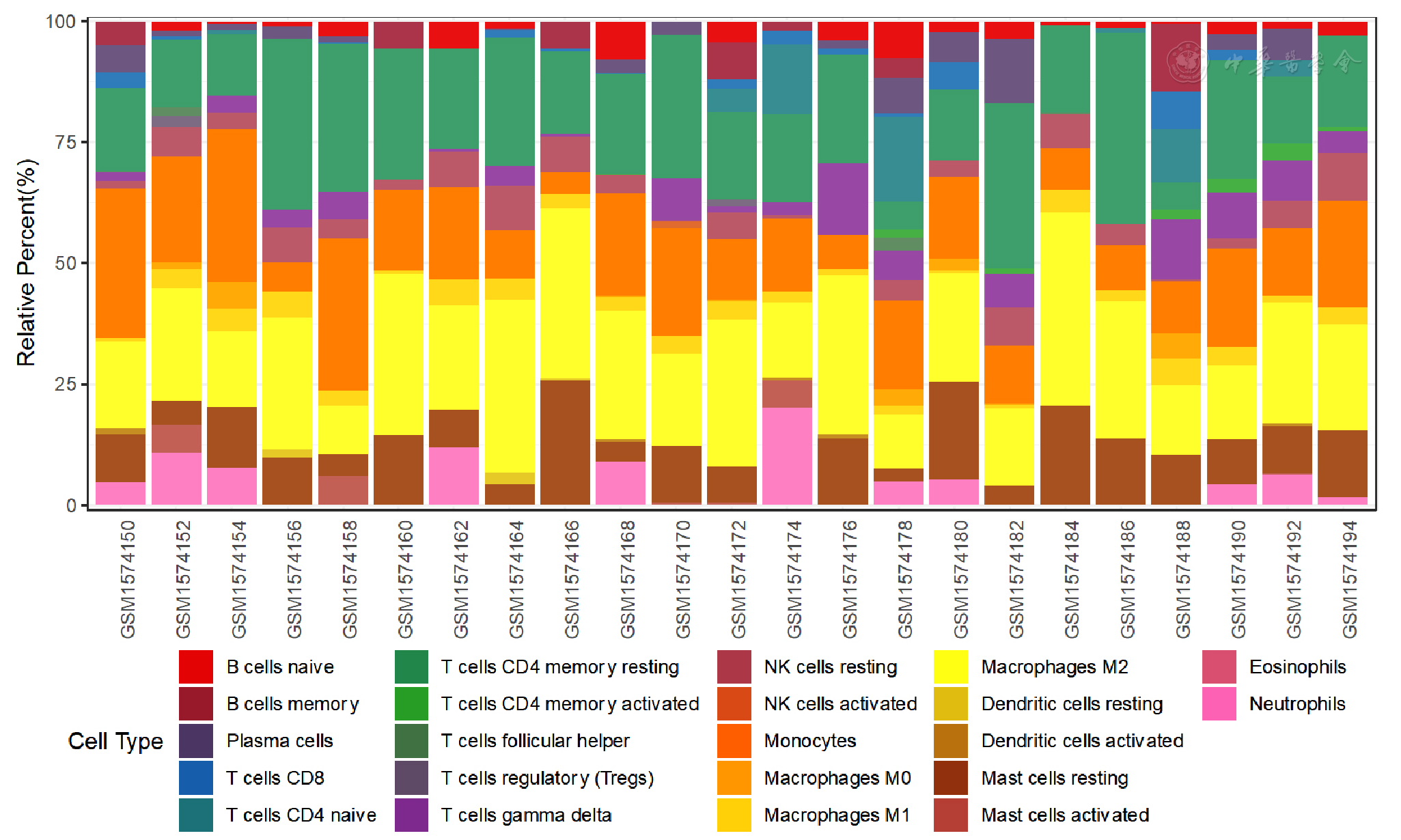
图12 GSE64554数据集EAT组织免疫细胞浸润柱状图
Figure 12 Bar graph of immune cell infiltration in epicardial adipose tissue between CAD patients and controls included in dataset GSE64554
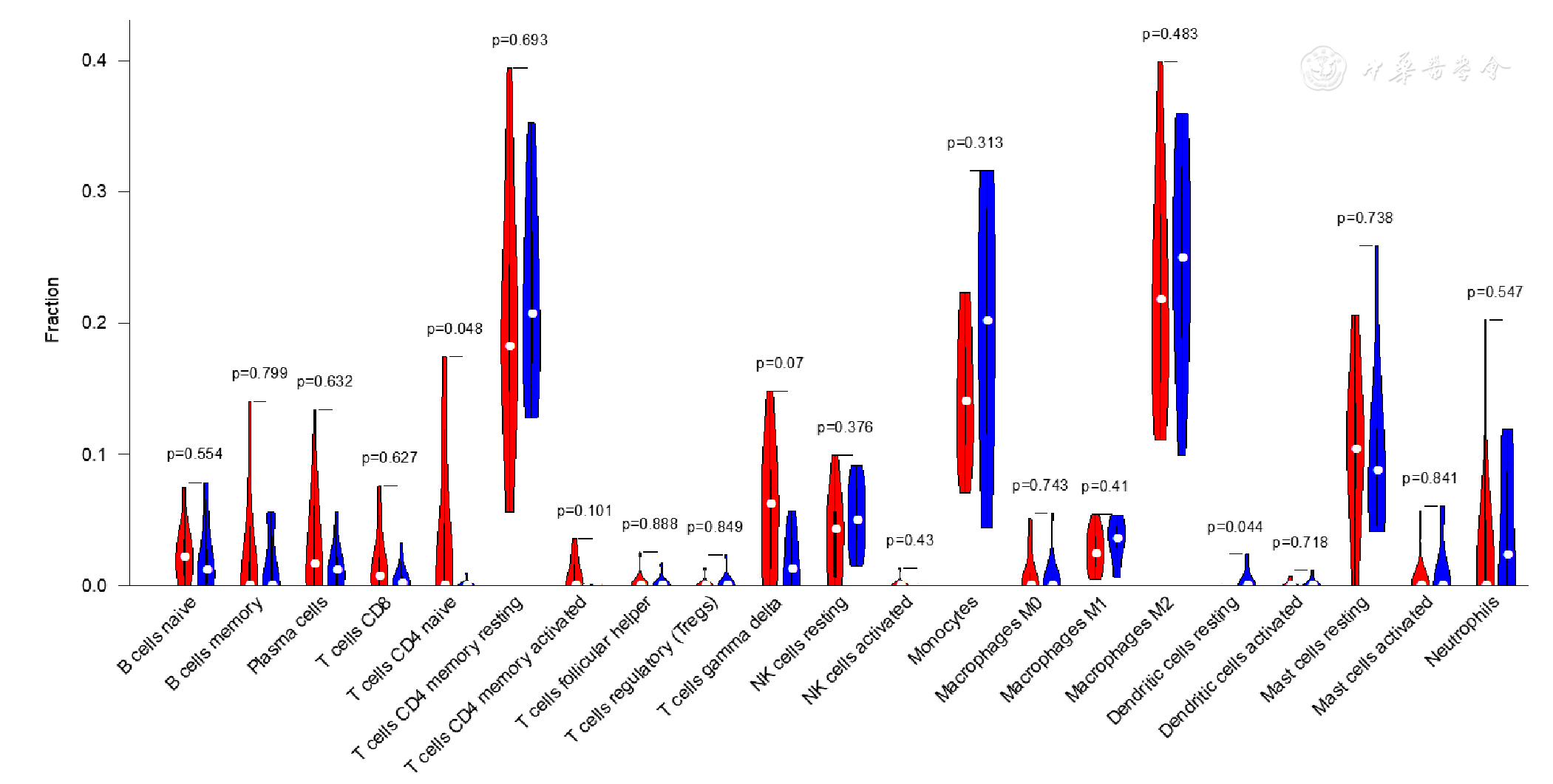
图13 GSE64554数据集EAT组织免疫细胞浸润小提琴图注:红色为CAD组,蓝色为健康对照组
Figure 13 Violin plot of immune cell infiltration in epicardial adipose tissue between CAD patients and controls included in dataset GSE64554
| 组别 | 样本数 | BPI | BIRC5 | CXCL12 | RNASE1 | F2R |
|---|---|---|---|---|---|---|
| CAD组 | 10 | 2.14±0.32 | 1.98±0.23 | 2.91±0.89 | 4.47±1.56 | 0.45±0.06 |
| 健康对照组 | 10 | 0.99±0.17 | 1.02±0.11 | 1.14±0.44 | 1.28±0.56 | 1.00±0.12 |
| t值 | -9.945 | -12.070 | -5.630 | -6.001 | 12.637 | |
| P值 | 0.011 | 0.010 | <0.001 | 0.009 | <0.001 |
表2 CAD组与健康对照组EXO差异性基因表达(±s)
Table 2 Differentially expressed genes in exosomes between CAD patients and healthy controls
| 组别 | 样本数 | BPI | BIRC5 | CXCL12 | RNASE1 | F2R |
|---|---|---|---|---|---|---|
| CAD组 | 10 | 2.14±0.32 | 1.98±0.23 | 2.91±0.89 | 4.47±1.56 | 0.45±0.06 |
| 健康对照组 | 10 | 0.99±0.17 | 1.02±0.11 | 1.14±0.44 | 1.28±0.56 | 1.00±0.12 |
| t值 | -9.945 | -12.070 | -5.630 | -6.001 | 12.637 | |
| P值 | 0.011 | 0.010 | <0.001 | 0.009 | <0.001 |
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [1] | 周启保, 罗潇, 陈玲, 曹俊达, 李菊香, 徐劲松, 苏海. 收缩压昼夜变异率联合正常RR间期标准差对冠心病诊断价值的临床研究[J]. 中国全科医学, 2025, 28(30): 3760-3765. |
| [2] | 吴文俊, 卫靖靖, 李雪, 任红杰, 于瑞, 彭广操, 朱明军. 基于隐结构模型结合关联规则分析冠心病合并高血压的方药规律[J]. 中国全科医学, 2025, 28(30): 3787-3795. |
| [3] | 杨继, 张垚, 赵英强, 张秋月. 中医三级防控模式对冠心病与脑卒中患者的管理效能评价:一项单中心前瞻性队列研究[J]. 中国全科医学, 2025, 28(22): 2750-2761. |
| [4] | 贺婷, 李佳, 谭文彬. 循环系统疾病与继发性骨质疏松症的研究新进展[J]. 中国全科医学, 2025, 28(17): 2101-2112. |
| [5] | 绳菁煜, 刘凡凡, 马梅, 田霖, 刘雨桐, 刘凤敏, 高杉, 于春泉. 冠心病患者血尿素氮与血清白蛋白比值与颈动脉斑块的相关性研究[J]. 中国全科医学, 2025, 28(15): 1831-1839. |
| [6] | 宋洪娜, 许洪梅, 刘玉环, 王青龙, 汤云昭, 于翔. 冠心病合并糖尿病患者经皮冠状动脉介入治疗术后共病随访模式的构建及应用效果研究[J]. 中国全科医学, 2025, 28(14): 1737-1743. |
| [7] | 柴依依, 叶青芳, 林平, 李玲. 寒地城市冠心病患者运动认知风险综合征发生现状及影响因素研究[J]. 中国全科医学, 2025, 28(14): 1730-1736. |
| [8] | 岑妮秒, 韦云师, 黄丽娜, 吴标良. 外泌体微小RNA参与糖尿病足溃疡修复的研究进展[J]. 中国全科医学, 2025, 28(09): 1156-1160. |
| [9] | 贾硕, 程功, 关蕾, 冯盼盼, 许百灵, 方伟, 张骥. 单电子发射计算机断层成像技术在经皮冠状动脉支架植入术后患者冠脉微循环障碍中的诊断性能研究[J]. 中国全科医学, 2025, 28(09): 1122-1127. |
| [10] | 李梦元, 李冠杉, 韩倩, 李思瑶, 郑舒畅, 辛明蔚, 尹晓丹, 王景尚, 武颖, 何军琴. 外泌体调节复发性流产小鼠子宫内膜细胞上皮间充质转化的作用机制研究[J]. 中国全科医学, 2025, 28(08): 962-972. |
| [11] | 岳海涛, 何婵婵, 成羽攸, 张森诚, 吴悠, 马晶. 基于机器学习的冠心病风险预测模型构建与比较[J]. 中国全科医学, 2025, 28(04): 499-509. |
| [12] | 杨红, 刘成, 刘森, 邵琪琪, 夭元昊, 付真彦. 残余胆固醇与进展为主要不良心血管事件的非罪犯病变易损斑块的相关性研究[J]. 中国全科医学, 2025, 28(03): 299-304. |
| [13] | 张铭, 王文娟, 郝问, 陈吉彬, 夏伟, 邵一兵, 王宾. 阻塞性睡眠呼吸暂停与急性冠脉综合征的相关性:临床研究的现状及展望[J]. 中国全科医学, 2025, 28(03): 257-261. |
| [14] | 孙雪纯, 杜智勇, 于华惠, 吕倩雯, 焦晓璐, 王钰, 秦彦文. 溶血磷脂类代谢物对急性冠脉综合征患者经皮冠状动脉介入治疗术后主要不良心血管事件的预测价值:一项前瞻性队列研究[J]. 中国全科医学, 2024, 27(36): 4540-4545. |
| [15] | 贾佳, 刘嘉慧, 季文君, 郑博, 王新刚, 范芳芳, 李寅, 张龙, 张岩. 急性心肌梗死患者血浆前蛋白转化酶枯草溶菌素9水平的影响因素研究[J]. 中国全科医学, 2024, 27(36): 4568-4574. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||





